Warning
This document is for an old release of Galaxy. You can alternatively view this page in the latest release if it exists or view the top of the latest release's documentation.
May 2022 Galaxy Release (v 22.05)

Highlights
The New History is Here
After years of effort by the development team and community contributors, the new history is finally here. It’s a big improvement over the old history interface, both technically and in terms of user experience, but it might take some getting used to. It has a lot of great new features like:
A quick switcher, letting you switch between recent histories
Easily find dataset inputs
Improved history search interface
Check it out now!
Storage Dashboard (beta)
Have you ever been over your quota? 250 GB sure goes fast! Now with the new Storage Dashboard you can get a quick overview of ways to help clean up deleted datasets and recover some of that quota allocation. This is a beta version of this interface, let us know if you have any issues. In future versions, it will be expanded to help you understand how your allocation is being used, and which histories and datasets should be cleaned up first.
Bulk History Operations
The new history allows for improved bulk operations, selecting dozens or hundreds of datasets to retag or change database key automatically, a process that used to require using collections or doing them one by one.
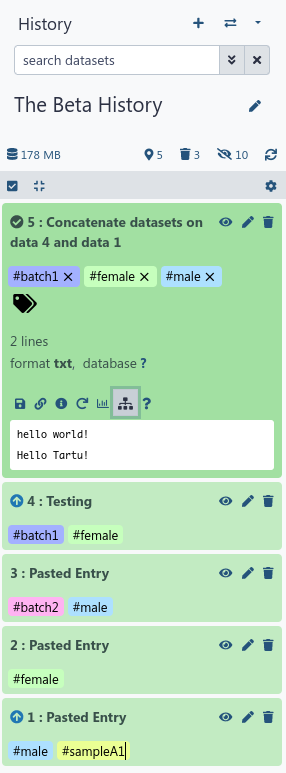
Deferred dataset resolution enables running workflows and tools without importing data into Galaxy first
It is now possible to defer dataset resolution for datasets imported via URLs and the “Choose remote files” dialog. This means that the deferred dataset will only be downloaded as it is needed during job execution. Deferred datasets will not count towards your storage quota, as the data is not stored by Galaxy. To enable deferred dataset resolution click on “Settings” in the upload dialog.
Workflow Improvements
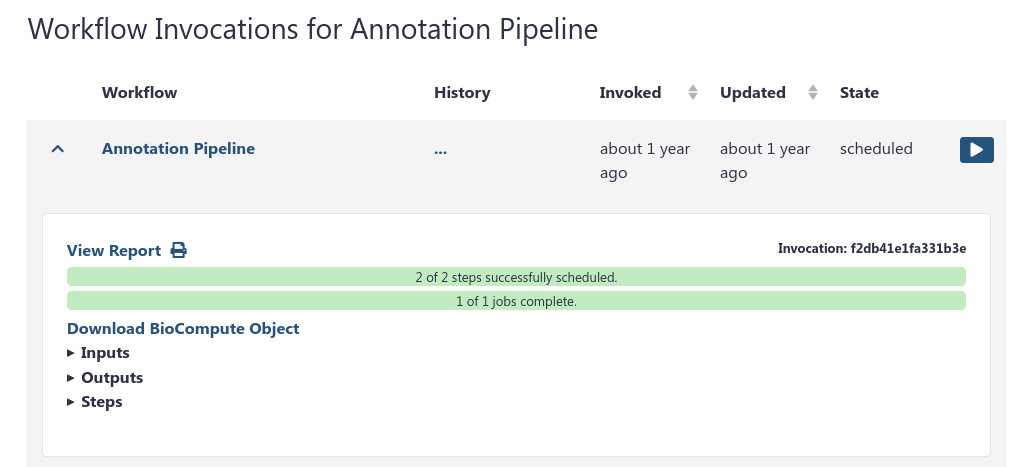
You can now see all invocations of a specific workflow, across all of your histories.
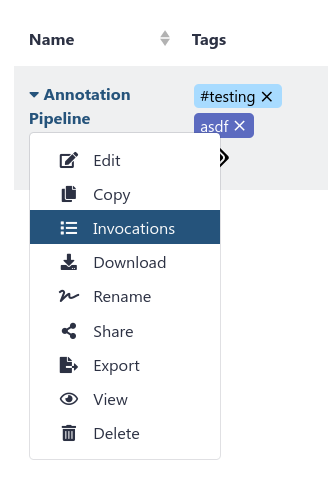
This is especially useful if you run a single workflow across multiple datasets, and keep them nicely separated in multiple histories. You can see all of the results in a single centralised location.
Additional workflow improvements have been made, e.g. numbering of steps in a workflow. Improvements have been made to the internal representation of the workflow that should make the saved .ga and .gxwf.yml files easier to compare across changes.
Workflows can also now be zoomed in and out via scrolling, this interface should be more familiar for anyone who has used Google Maps:
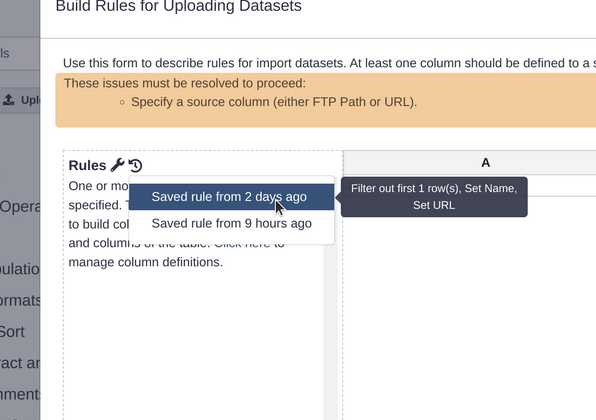
Saved Rules
Do you use the Rule Based Uploader (RBU)? (If not, learn how today!) The RBU now has a very convenient recently used rule listing, sorted by how recently they were used. You can hover over each entry to also list a preview of what steps were done in that RBU invocation:
Scratchbook Upgraded
The scratchbook has been updated to a new implementation. Now your views of datasets can overlap, you have much more freedom in the size of each window, and you can minimize datasets like minimizing a window to the taskbar in Windows.
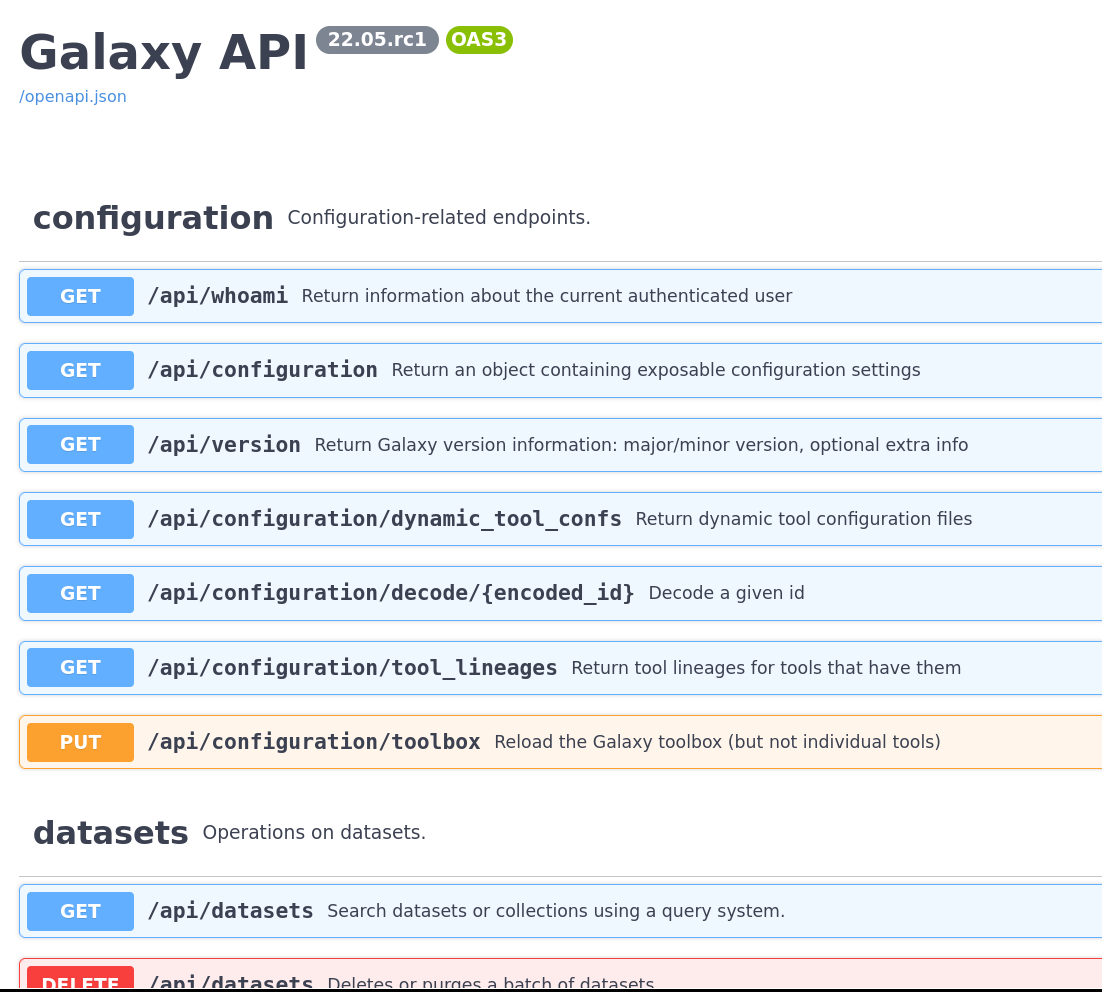
OpenAPI Docs
The Galaxy API is going through an upgrade to FastAPI, which allows us to generate OpenAPI documentation. If you’re an API consumer, either via BioBlend or via another system, you can use this to see all of the APIs available from Galaxy and try them out live.
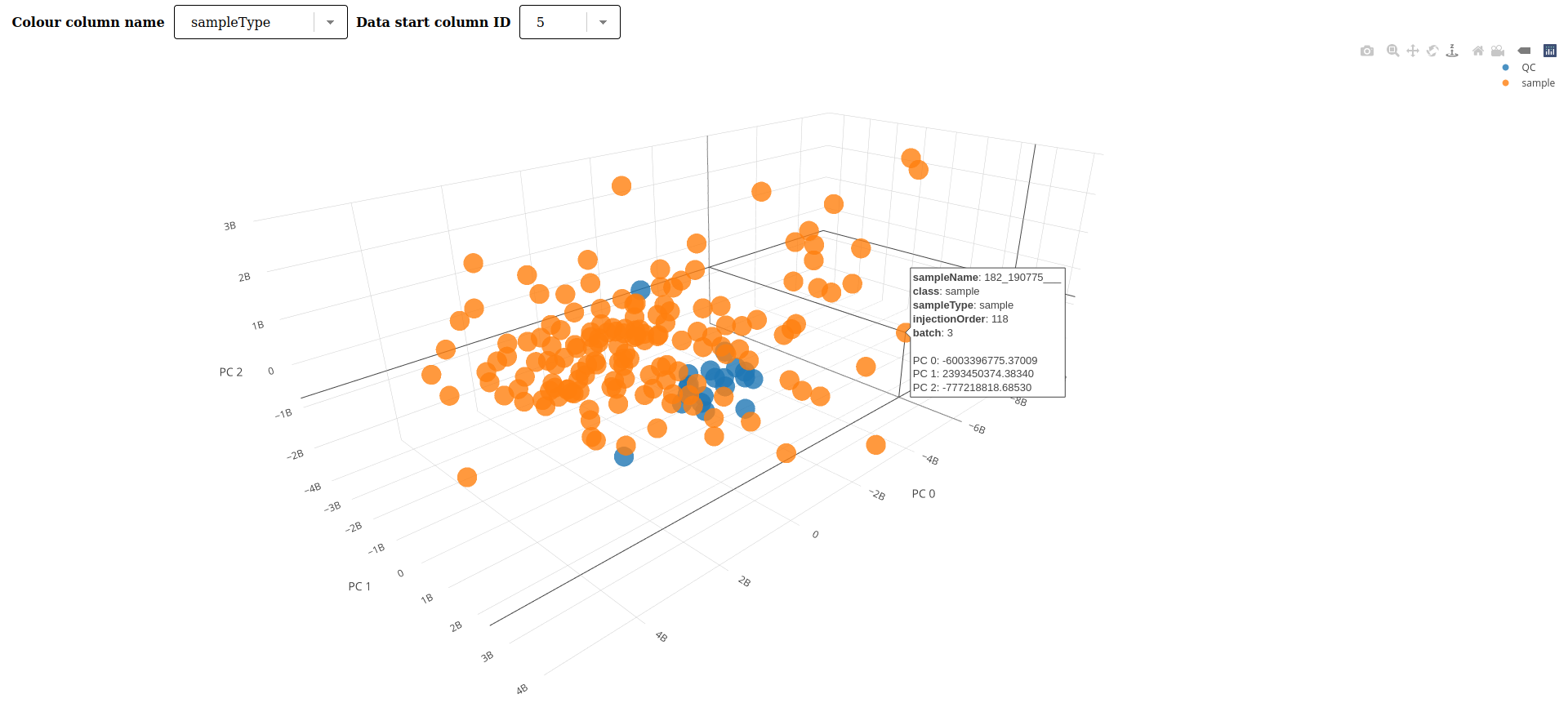
New Visualizations
Add gulp target for force-rebuild of viz plugins (thanks to @dannon). Pull Request 13370
Properly initialize chart settings for new visualizations (thanks to @guerler). Pull Request 13561
Point to GTN tutorial in visualizations readme (thanks to @beatrizserrano). Pull Request 13616
Add 3D PCA visualization (thanks to @xtrojak). Pull Request 13771
Support phylocanvas autobuild (thanks to @dannon). Pull Request 14023
Fix a bug in PCA artifact staging that prevents a plugin rebuild (thanks to @dannon). Pull Request 14148
Visualization fix for MSA (thanks to @dannon). Pull Request 14263
Add 3D PCA visualization (thanks to @xtrojak). Pull Request 13771
New Datatypes
Python code formatting standard, automatic (thanks to @dannon). Pull Request 11162
Add new molecule datatypes (thanks to @muon-spectroscopy-computational-project). Pull Request 12375
Upgrade syntax to Python 3.7 (thanks to @nsoranzo). Pull Request 13500
Add mzqc datatype (thanks to @bernt-matthias). Pull Request 13564
Update mypy to 0.942 and fix new mypy errors (thanks to @nsoranzo). Pull Request 13650
Create data_fetch and set_metadata celery tasks (thanks to @mvdbeek). Pull Request 13655
Add bigWig to Wig converter (thanks to @gallardoalba). Pull Request 13708
Replace last use of deprecated
impmodule (thanks to @nsoranzo). Pull Request 13817Add support for gz-compressed gfa1 and gfa2 files (thanks to @gallardoalba). Pull Request 13927
Tighten sniffers (thanks to @mvdbeek). Pull Request 13959
Don’t create a BGZF index when running the compressing converter (thanks to @nsoranzo). Pull Request 13969
Fix bug: set creation_date on dataset.metadata, not dataset (thanks to @jdavcs). Pull Request 14039
Fix composite datatypes with
substitute_name_with_metadata(thanks to @mvdbeek). Pull Request 14053Enable generating urls for WSGI routes within FastAPI routes (thanks to @mvdbeek). Pull Request 14066
Fix collection download for BAM native files (thanks to @mvdbeek). Pull Request 14162
New Datatype: QIIME 2 (thanks to @ebolyen). Pull Request 14216
Make Agp set_meta more lenient (thanks to @astrovsky01). Pull Request 14277
Deleted, discarded and deferred file not found tweaks (thanks to @mvdbeek). Pull Request 14296
Add scool datatype (thanks to @bernt-matthias). Pull Request 14480
Fix xml deprecation warning in
parse_requirements_from_xml(thanks to @bernt-matthias). Pull Request 14483Add new molecule datatypes (thanks to @elichad). Pull Request 12375
Add mzqc datatype (thanks to @bernt-matthias). Pull Request 13564
Add bigWig to Wig converter (thanks to @gallardoalba). Pull Request 13708
Add support for gz-compressed gfa1 and gfa2 files (thanks to @gallardoalba). Pull Request 13927
Fix collection download for bam native files (thanks to @mvdbeek). Pull Request 14162
Builtin Tool Updates
Python code formatting standard, automatic (thanks to @dannon). Pull Request 11162
Add built-in converters to tool panel (thanks to @jdavcs). Pull Request 13447
Add JupyTool, user-configurable interactive Jupyter-based tool (thanks to @blankenberg). Pull Request 13606
Fix David URL (thanks to @hexylena). Pull Request 13666
Add and update various interactive tools (thanks to @gmauro). Pull Request 13811
Bump ncb-datasets-cli dependency to 13.14.0 (thanks to @mvdbeek). Pull Request 13853
New Datatype: QIIME 2 (thanks to @ebolyen). Pull Request 14216
Fixes for JupyTool, and overlapping versions with jupyter (thanks to @blankenberg). Pull Request 14309
remove debug statements from tool search (thanks to @bgruening). Pull Request 14386
Grep1 tool: fix keep_header option (thanks to @jancrichter). Pull Request 14475
Overhaul and improve the Jupyter Interactive Tool, providing many new interface options (thanks to @blankenberg). Pull Request 13606 * You can specify any number of user-defined inputs using the repeat input, providing name value, selecting the type of input, and then providing values. * You can make the JupyTool reusable in a workflow, by allowing the user to specify input values for the defined input blocks. * Inputs can be accessed by name from the automatically provided GALAXY_INPUTS dictionary. * Outputs can be written automatically to the user’s history by writing to the outputs directory for one individual file or to the outputs/collection directory for multiple files. * Using collection tools, you can parse out the individual elements from the collection, as needed.
Add built-in converters to tool panel (thanks to @jdavcs). Pull Request 13447
Add and update various interactive tools (thanks to @gmauro). Pull Request 13811
Bump ncb-datasets-cli dependency to 13.14.0 (thanks to @mvdbeek). Pull Request 13853
Release Testing Team
A special thanks to the release testing team for testing many of the new features and reporting many bugs:
Ahmed Awan <https://github.com/ahmedhamidawan>
Catherine Bromhead <https://github.com/cat-bro>
Fabio Cumbo <https://github.com/cumbof>
Jennifer Hillman-Jackson <https://github.com/jennaj>
John Davis <https://github.com/jdavcs>
Simon Bray <https://github.com/simonbray>
Tyler Collins <https://github.com/tcollins2011>
Release Notes
User facing release notes compiled by Helena Rasche.
Please see the full release notes for more details.
To stay up to date with Galaxy’s progress, watch our screencasts; visit our community Hub; and follow us on Bluesky, Mastodon, and LinkedIn.
You can always chat with us on Matrix.
Thanks for using Galaxy!