Warning
This document is for an in-development version of Galaxy. You can alternatively view this page in the latest release if it exists or view the top of the latest release's documentation.
January 2020 Galaxy Release (v 20.01)

Highlights
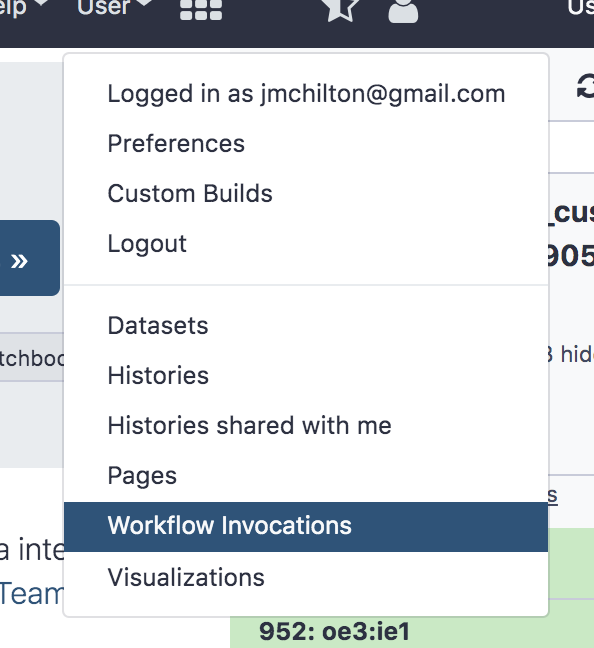
- Workflow Executions Menu
This menu can be reached from the User menu -> Workflow Invocations and lists your recent Workflow executions, their status and links to the Workflow Editor and the History with the results of the Workflow run.

- Galay Markdown Pages and Workflow Reports as PDF
An additional link has been added to Galaxy Markdown Pages and Workflow reports that exports your document as a standalone PDF.

- Screenreader-friendly Navigation
The top menu and the Workflow Editor have been improved for easier navigation using Screenreaders and the Keyboard. Thanks goes to @WilliamHolden
- Email-notification for completed jobs
You can now select to be notified by email when a tool run has finished. Thanks goes to @astrovsky01

New Visualizations
Add data dialog input element to client form parameter list Pull Request 8774
Wrap SQL statement in a text() function as required by sqlalchemy (thanks to @kaukrise). Pull Request 9216
Ensure visible overflow on navbar container (thanks to @kaukrise). Pull Request 9217
New Datatypes
Datatypes for MassSpec DIA dlib,elib (thanks to @jj-umn). Pull Request 8657
Add bus as binary, remove extra datatype (thanks to @astrovsky01). Pull Request 8705
remove dada2_derep data type (thanks to @bernt-matthias). Pull Request 8902
Add new GROMACS datatypes: ndx, xvg, edr (thanks to @simonbray). Pull Request 8964
add gff3.gz and gff3.bz2 datatypes (thanks to @FredericBGA). Pull Request 8967
Make mol2 visible (thanks to @galaxyproject). Pull Request 8995
Add a datatype subclass for CAT (Contig Annotation Tool) (thanks to @jj-umn). Pull Request 9087
Fix cool sniffing on python3 Pull Request 9153
Allow tabular.gz / add embl to uploadable formats Pull Request 9293
Builtin Tool Updates
Upgrade askomics interactive tool (thanks to @xgaia). Pull Request 8517
Remove collection ID character restriction from ‘Relabel collection from file’ tool (thanks to @brinkmanlab). Pull Request 8571
Allow space, dot and comma in relabel identifier tool Pull Request 8757
Fix script shebangs (thanks to @nsoranzo). Pull Request 8888
grouping: strip newlines (thanks to @bernt-matthias). Pull Request 8953
Allow admins to import tools/workflows from paths. Pull Request 9003
Add requirements to interval_to_bed_converter.xml Pull Request 9041
Move sam_pileup to GALAXY_LIB_TOOLS_VERSIONED Pull Request 9320
Add requirements, fix legacy tools and move them out of GALAXY_LIB_TOOLS_UNVERSIONED Pull Request 9336
Add requirements to (almost) all converters, profile and minor fixes Pull Request 9345
Fix version in which
sam_pileupwas fixed (thanks to @nsoranzo). Pull Request 9350Expunge tool shed repository objects, prevents ParentInstanceDetachedError Pull Request 9354
Remove GenomeSpace tools, OpenID, UI and requirements (thanks to @nsoranzo). Pull Request 9363
Fix maf tools Pull Request 9393
XML fix for joiner.xml. Pull Request 9399
Release Notes
Please see the full release notes for more details.
To stay up to date with Galaxy’s progress, watch our screencasts; visit our community Hub; and follow us on Bluesky, Mastodon, and LinkedIn.
You can always chat with us on Matrix.
Thanks for using Galaxy!