25.0 Galaxy Release (June 2025)

Please see the full 25.0 release notes for more details.
Highlights
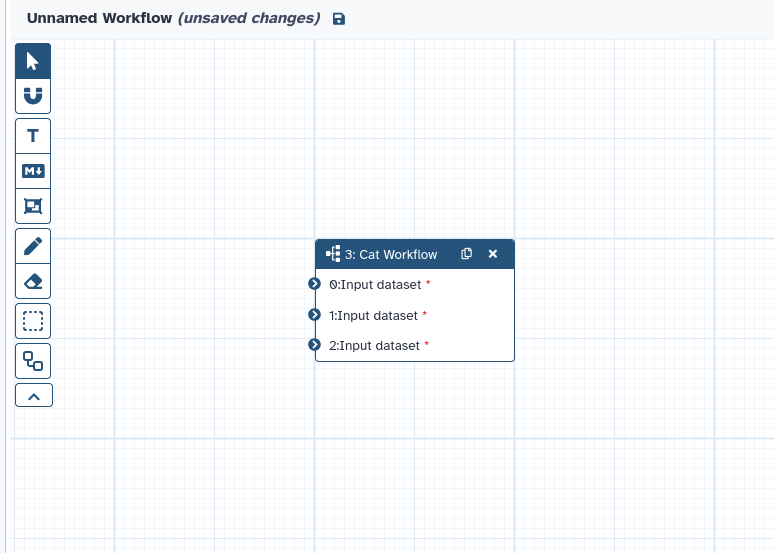
Empower Users to Build More Kinds of Collections, More Intelligently
Galaxy’s collection system has been vastly expanded, simplifying creation while supporting new, advanced use cases for both users and developers.
Support for mixed paired and unpaired collections. Tools and workflows can now work with lists that contain both paired and unpaired data (list:paired_or_unpaired), supporting more flexible scientific analyses and tool development. A new collection operation tool lets you split such collections into homogeneous lists if needed.
Generalized collection creation. Easily build these new generalized collection types—including nested lists (list:list), nested lists of pairs (list:list:paired), and more—directly from selected datasets, without requiring rules.
Wizard-style collection builders. A new wizard component guides users step-by-step, auto-detecting data structures and suggesting the most appropriate collection format. Choose between “Auto Build List” for instant setup or “Advanced Build List” for full manual control, with tailored help and warnings.
New Rule Based Import Activity. An activity-based wizard for rule-based uploads gives more space, clarity, and flexibility. Notably, you can now drop files directly onto the “Paste Table” form, and work with rules in a more consistent, central location.
Better error handling and feedback. The new interfaces provide clear feedback and tailored warnings for duplicate identifiers and unpaired datasets, improving the user experience.
Refinements and bug fixes. Includes fixes for extension handling, improved abstractions for collection input adaptation, and refactoring for easier future maintenance.
These changes make collection creation more powerful and accessible, supporting both simple and sophisticated data organization scenarios, and laying the groundwork for future enhancements. [#19377]
Here’s an example of one of the new collection builders in action:
Add ZIP explorer to import individual files from local or remote ZIP archives
Galaxy now features a ZIP explorer, refining the process of importing individual files from both local and remote ZIP archives.
ZIP explorer integration. Browse the contents of ZIP files directly in Galaxy and select only the files you need, minimizing unnecessary uploads.
Support for local and remote archives. Import from ZIP files on your computer or by providing a remote URL.
Improved user experience. The intuitive interface simplifies file selection and import, making it easier to work with compressed data. It also recognizes some known archive structures like RO-Crate and Galaxy export archives, and will display additional useful information in those cases.
These enhancements make importing data from ZIP archives more flexible and efficient, further reducing barriers to working with large or complex datasets. [#20054]
A New Unified View for Datasets
Galaxy 25.0 introduces a unified view for datasets, consolidating common dataset actions into a single, streamlined interface.
Single access point. Instead of multiple separate buttons, users now interact with a single button on each dataset, which opens a tabbed interface for all dataset-related actions.
Tabbed layout. Key functionality — such as viewing metadata, peeking into file contents, accessing datatypes/permissions and more — is now organized into clear, intuitive tabs.
Consolidated interface. This update provides a one stop shop for dataset management, reducing clutter and confusion from multiple buttons and options.
This change enhances consistency across the Galaxy interface and offers a more intuitive experience for both new and experienced users working with datasets. [#20154]
Allow Rerunning Workflows with the Same Inputs and Parameters
Users can now easily rerun any workflow with the exact same inputs and parameters as a previous run - with a simple click of the new “Rerun” button in the workflow invocation view. This feature simplifies reproducibility and troubleshooting by letting you repeat analyses with a single click. The workflow run form also detects and notifies of any changes to inputs compared to the original run! [#20032]
An Enhanced Workflow Run Form Interface
Galaxy 25.0 introduces a streamlined workflow run interface that allows users to upload inputs, create collections, and configure parameters — all within a single, unified form.
Integrated data upload and collection creation. Users can now upload datasets or build collections directly from the workflow run form, without navigating away or switching contexts.
Visual input indicators. Each input now displays a visual status indicator, making it easy to see which inputs are ready and which still need attention.
Improved settings panel. The run form includes a clearer, more organized settings menu with well-annotated options for configuring the workflow execution.
Live-updating workflow graph. A dynamic graph of the workflow appears alongside the form, updating in real time as users populate inputs — helping them understand the structure and flow of the analysis.
These improvements were inspired by users encountering curated workflows from platforms like the IWC Browser or BRC Analytics , and are designed to reduce friction, improve clarity, and enable users to complete complex analyses entirely within a single interface. [#19294]
Introducing READMEs for Workflows in the Galaxy Interface
Galaxy 25.0 introduces support for associating README/help text with workflows. This provides a way to document workflows directly in the Galaxy interface, with support for Markdown formatting, links, and images.
README field for workflows. Each workflow can now have an associated README string, stored as part of the workflow object but separate from the workflow steps or structure.
Viewable on the run form. A new “Show Workflow Help” button on the workflow run form reveals associated documentation alongside input parameters — helping users understand how to use a workflow.
Editable from the workflow editor. Workflow authors can scroll the “Attributes” panel in the editor, click “Show Readme,” and add or update the associated documentation.
This feature lowers the barrier to understanding and reusing workflows, especially in collaborative environments. [#19591]
Bulk Actions for Workflows
Galaxy 25.0 introduces support for selecting multiple workflows to perform actions in bulk, simplifying workflow management for users.
Selectable workflow cards. In the “My Workflows” tab, users can now select multiple workflows at once.
Bulk operations. Perform common actions — such as delete, restore, and tag — on all selected workflows simultaneously.
Improved tagging interface. A new tag selection dialog makes it easier to organize workflows with consistent tagging.
These enhancements simplify workflow housekeeping and make it easier to manage shared or long-running projects involving many workflows. [#19336]
Redesigned File Sources and Storage Location Interfaces
Galaxy 25.0 introduces a modernized interface for accessing Remote File Sources and Storage Locations, making navigation and selection more intuitive and user-friendly.
Card-based layout. Available options are now displayed in a clear, responsive card view for improved readability and usability.
Breadcrumb navigation. A breadcrumb bar helps users track their position and move between sections more easily.
Search and filter support. A built-in search field allows users to quickly locate file sources or storage locations by name.
These changes provide a cleaner and more consistent interface for interacting with file sources and storage locations. [#19521]
Enable Cloning Subworkflows in the Workflow Editor
In the Workflow Editor, subworkflows can now be cloned just like regular steps. This allows users to easily duplicate subworkflows within a larger workflow, with a singular click of the “Duplicate” button on a subworkflow node. [#19420]
JupyterLite for Lightweight In-Browser Notebooks within Galaxy
JupyterLite — a fully browser-based Jupyter environment — is now available as a visualization (accessible right in the Galaxy center panel) with support for interacting directly with Galaxy datasets and APIs, without requiring any local setup, or using the Jupyter interactive tool.
JupyterLite integration. Launch Python notebooks directly in the visualization using the jl-galaxy extension.
Access Galaxy data. Use the gxy utility module to download datasets (await gxy.get()), upload results (await gxy.put()), or access the Galaxy API (await gxy.api()).
No setup required. No need for Conda, Docker, or Python installations — just open a JupyterLite notebook visualization in Galaxy, and start coding.
This makes interactive data exploration and scripting more accessible, where the user prefers to stay within the Galaxy interface without needing to switch to a separate Jupyter interactive tool. [#20174]
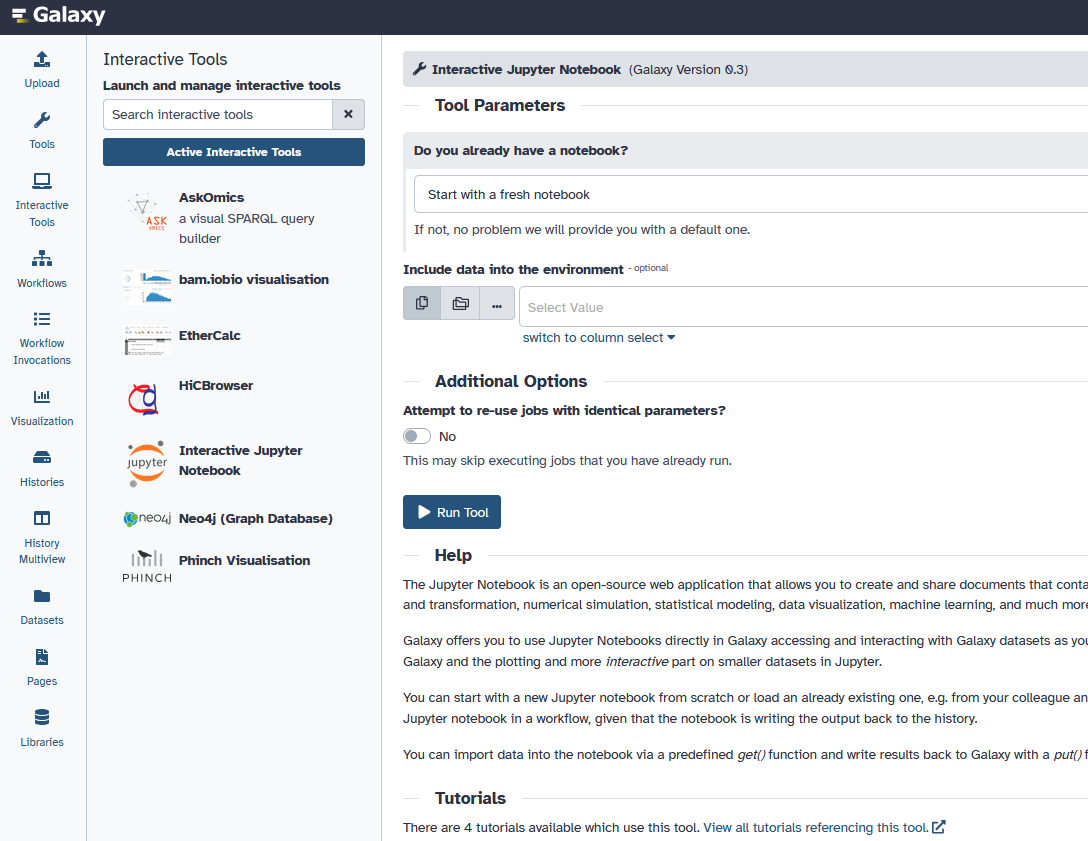
New Interactive Tools Panel in the Activity Bar
Interactive Tools (ITs) in Galaxy now have their own dedicated panel in the Activity Bar, providing a clearer, more accessible experience for users launching and managing interactive environments.
Separate from standard tools. Previously nested under a section in the main Tools panel, Interactive Tools are now listed under their own Activity Bar activity — making them easier to find.
Visual icons for each tool. Each IT is represented with its own icon for faster recognition.
Live status indicators. A badge on the Activity icon shows how many ITs are currently running.
This update helps distinguish Interactive Tools from standard tools, improves discoverability, and provides a more intuitive way to monitor and manage interactive sessions. [#19996]
Preferred Visualization Display for Dataset Types
Galaxy now allows administrators to configure default visualizations for specific datatypes. Instead of showing raw file content in the (new) Preview tab, datasets with a preferred visualization will now display that visualization by default.
This improves the default experience for formats like HDF5, where binary previews are not useful, and makes interactive visualizations immediately accessible to users. [#20190]
Here is a quick example:
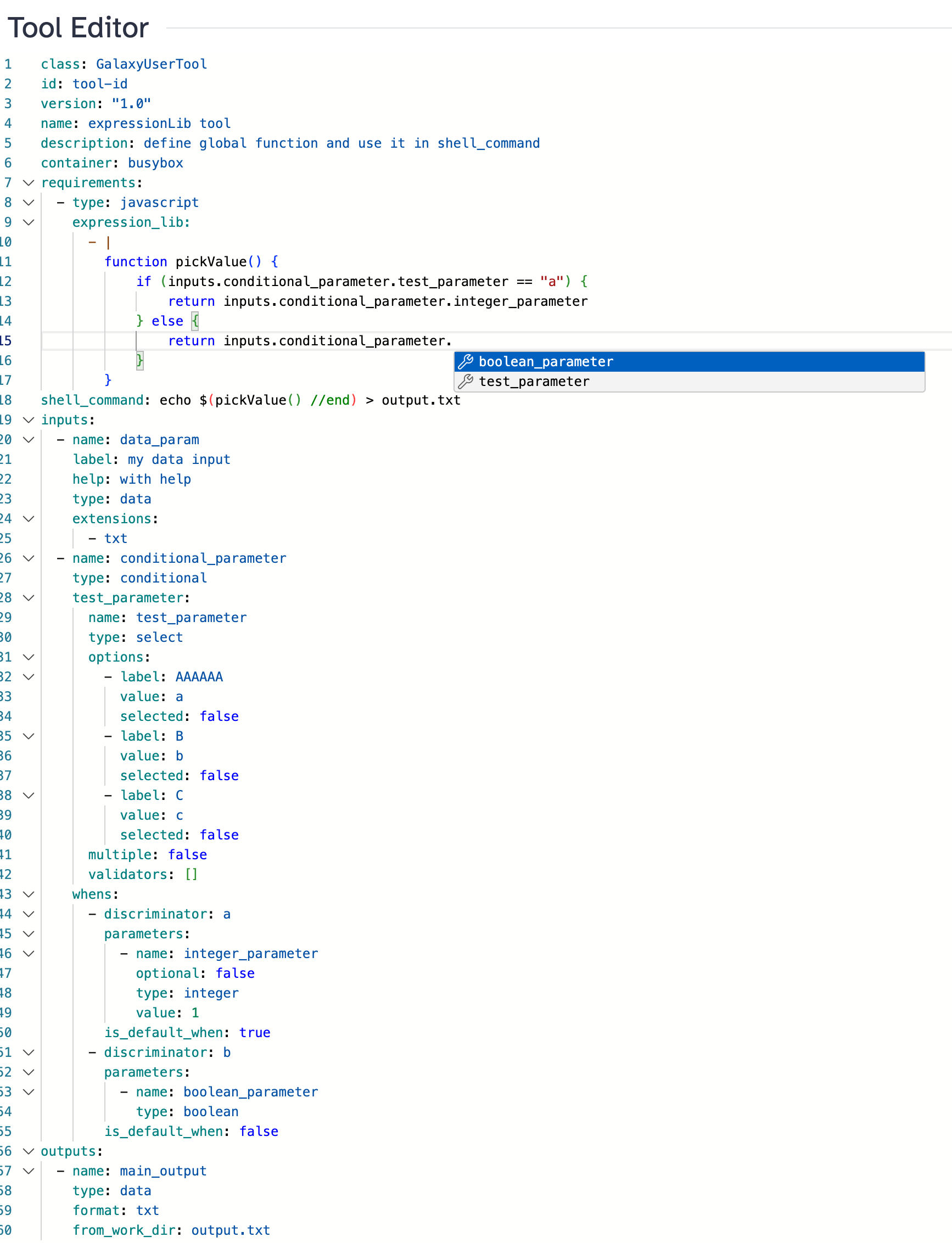
User-Defined Tools (Beta)
Galaxy 25.0 introduces support for user-defined tools — a new feature that lets selected users define and run tools directly in the Galaxy interface, without requiring regular administrative intervention.
Create and run tools without admin requests. For the first time, users can define tools without needing to go through the standard installation process.
YAML-based tool definitions. Tools are written in a structured YAML format and stored in the Galaxy database. They include inputs, outputs, and shell commands written using sandboxed JavaScript expressions.
Container execution only. All user-defined tools must run inside containers for enhanced security.
Workflow support. User-defined tools are automatically included when shared as part of a workflow, ensuring seamless collaboration.
This feature is currently in beta and must be explicitly enabled by administrators. It is recommended only for trusted users while safety audits are ongoing. [#19434]
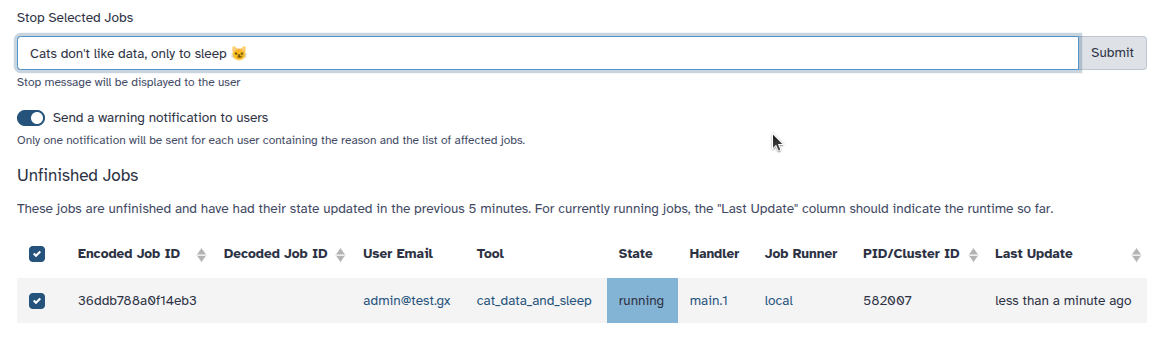
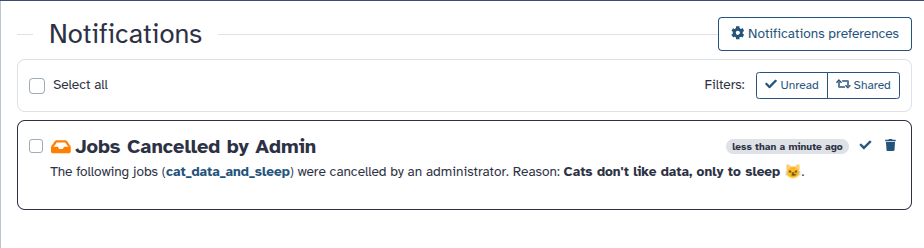
Job Cancellation Notifications for Admins
Administrators can now notify users when their jobs are canceled, improving transparency and communication.
Optional user notification. When canceling jobs, admins can choose to send a notification message to affected users.
Grouped notifications. Users receive a single notification listing all canceled jobs, even if multiple jobs were terminated at once.
Job details view. Clicking on a listed job in the notification opens a detailed job summary page.
Supports email delivery. If the user has email notifications enabled, they will also receive the message by email.
This feature gives administrators more control over user communication during job management actions. [#19547]
eLabFTW integration
Galaxy is now integrated with eLabFTW, a free and open source electronic lab notebook from Deltablot via a new file source.
eLabFTW can keep track of experiments, equipment and materials from a research lab. Each lab can either host their own installation or go for Deltablot’s hosted solution. A live demo showcasing its features is available here.
Files attached to eLabFTW experiments and resources can be imported to histories with just a few clicks. After the analysis is complete, datasets and histories can be exported back as attachments to an experiment or resource.
Further details are available on this blog post.
RSpace integration
Galaxy 25.0 includes a new file source that integrates it with RSpace, an open-source research data management system (RDM) and electronic lab notebook (ELN) providing inventory and sample management systems and featuring a wide collection of integrations with other services.
Thanks to this integration, files from the RSpace Gallery can be easily imported to Galaxy histories. After the analysis is complete, datasets and even whole histories can be exported back to the Gallery.
Read more on this blog post.
Visualizations
Update tiffviewer to latest version supporting more formats (thanks to @davelopez). Pull Request 20457
Rename vitessce_json file_ext to vitessce.json (thanks to @mvdbeek). Pull Request 20473
Add
/api/datasets/{dataset_id}/extra_files/raw/{filename:path}(thanks to @mvdbeek). Pull Request 20468Remove legacy visualizations (thanks to @guerler). Pull Request 20173
Fix phylocanvas visualization build (thanks to @davelopez). Pull Request 19138
Add Vizarr visualization (thanks to @davelopez). Pull Request 19061
Move phylocanvas to script entry point (thanks to @guerler). Pull Request 19193
Move heatmap visualization to new script endpoint (thanks to @guerler). Pull Request 19176
Add plotly.js (thanks to @guerler). Pull Request 19206
Switch h5web to script endpoint (thanks to @guerler). Pull Request 19211
Update visualizations to latest charts package (thanks to @guerler). Pull Request 19213
Update Vizarr package version to 0.1.6 (thanks to @davelopez). Pull Request 19228
Allow embedding vitessce visualizations (thanks to @mvdbeek). Pull Request 19909
FITS Graph Viewer - script name tweak (thanks to @dannon). Pull Request 19902
Update vitessce version (thanks to @mvdbeek). Pull Request 20016
Add kepler.gl visualization (thanks to @guerler). Pull Request 20005
Add Niivue viewer (thanks to @guerler). Pull Request 19995
Add VTK Visualization Toolkit Plugin (thanks to @guerler). Pull Request 20028
Add support for Markdown help text in visualizations (thanks to @guerler). Pull Request 20043
Add sample datasets for visualizations (thanks to @guerler). Pull Request 20046
Adds Example Datasets and Help Text for Visualizations (thanks to @guerler). Pull Request 20097
Add Molstar (thanks to @guerler). Pull Request 20101
Add alignment.js for multiple sequence alignment rendering (thanks to @guerler). Pull Request 20110
Add logo, description and help for aequatus (thanks to @guerler). Pull Request 20128
Migrate Transition Systems Visualization (thanks to @guerler). Pull Request 20125
Fix and migrate Drawrna (thanks to @guerler). Pull Request 20102
Add updated PCA plot (thanks to @guerler). Pull Request 20140
Hide non-functional and replaced visualizations (e.g. Nora, MSA) (thanks to @guerler). Pull Request 20077
Restore Visualization insertion options in Reports Editor (thanks to @guerler). Pull Request 20000
Add visualization test data (thanks to @nilchia). Pull Request 20183
Add plotly 6.0.1 to JupyterLite (thanks to @guerler). Pull Request 20201
Browse multiple trees in phylocanvas (thanks to @guerler). Pull Request 20141
Add Vitessce Viewer (thanks to @guerler). Pull Request 19227
Add JupyterLite (thanks to @guerler). Pull Request 20174
Migrate ChiraViz (thanks to @guerler). Pull Request 20214
Enable visualizations for anonymous user (thanks to @guerler). Pull Request 20210
Remove backbone-based charts modules (thanks to @guerler). Pull Request 19892
Improve handling of very large files in Tabulator (thanks to @guerler). Pull Request 20271
Datatypes
Add Aladin as standard FITS viewer (thanks to @bgruening). Pull Request 20465
Dataset Display and Preferred Viz fixes (thanks to @dannon). Pull Request 20439
Make optional edam-ontology in datatypes registry optional (thanks to @natefoo). Pull Request 20492
Rename vitessce_json file_ext to vitessce.json (thanks to @mvdbeek). Pull Request 20473
Add molstar as default viewer for some molecule formats (thanks to @bgruening). Pull Request 20467
Handle isatools dependency (thanks to @jdavcs). Pull Request 19582
Enhance UTF-8 support for filename handling in downloads (thanks to @arash77). Pull Request 19161
Calculate hash for new non-deferred datasets when finishing a job (thanks to @nsoranzo). Pull Request 19181
Fix UP031 errors - Part 4 (thanks to @nsoranzo). Pull Request 19235
Fix UP031 errors - Part 5 (thanks to @nsoranzo). Pull Request 19282
Fix UP031 errors - Part 6 (thanks to @nsoranzo). Pull Request 19314
Type annotation fixes for mypy 1.14.0 (thanks to @nsoranzo). Pull Request 19372
Add IGB display support for CRAM files (thanks to @paige-kulzer). Pull Request 19428
Format code with black 25.1.0 (thanks to @nsoranzo). Pull Request 19625
Fail request explicitly when sqlite provider used on non-sqlite file (thanks to @mvdbeek). Pull Request 19630
Add fastk_ktab_tar datatype required for fastk tool (thanks to @SaimMomin12). Pull Request 19615
Raise
MessageExceptionwhen using data provider on incompatible data (thanks to @mvdbeek). Pull Request 19639Drop support for Python 3.8 (thanks to @nsoranzo). Pull Request 19685
Use model classes from
galaxy.modelinstead ofapp.modelobject - Part 2 (thanks to @nsoranzo). Pull Request 19726Add bwa_mem2_index directory datatype, framework enhancements for testing directories (thanks to @mvdbeek). Pull Request 19694
Fix anndata metadata setting for data with integer indexes (thanks to @mvdbeek). Pull Request 19774
Add rDock prm datatype (thanks to @nsoranzo). Pull Request 19783
Fix parameter model constructions with leading underscores, fixes converter linting (thanks to @mvdbeek). Pull Request 19790
Bump up max_peek_size to 50MB (thanks to @mvdbeek). Pull Request 19823
Fix tabular metadata setting on pulsar with remote metadata (thanks to @mvdbeek). Pull Request 19891
Populate image metadata without allocating memory for the entire image content (thanks to @kostrykin). Pull Request 19830
Let pysam use extra threads available in job (thanks to @mvdbeek). Pull Request 19917
Extend image metadata (thanks to @kostrykin). Pull Request 18951
Add vitesscejson datatype (thanks to @guerler). Pull Request 20027
Add new line to vtpascii test file (thanks to @guerler). Pull Request 20051
Add flac audio format (thanks to @bgruening). Pull Request 20057
Add specific datatypes for Cytoscape and Kepler.gl (thanks to @guerler). Pull Request 20117
Add rd datatype (thanks to @richard-burhans). Pull Request 20060
Update tabular_csv.py to use less memory in tsv->csv conversion (thanks to @cat-bro). Pull Request 20187
Add more metadata, esp infer_from to datatypes configuration (thanks to @bgruening). Pull Request 20142
Add docx datatype (thanks to @bgruening). Pull Request 20055
Add markdown datatype (thanks to @bgruening). Pull Request 20056
Add bigbed to bed converter and tests (thanks to @d-callan). Pull Request 19787
Visualization-First Display functionality (thanks to @dannon). Pull Request 20190
Refactor display_as URL generation for UCSC links and fix to remove double slashes in URL (thanks to @natefoo). Pull Request 20239
Builtin Tool Updates
Format code with black 25.1.0 (thanks to @nsoranzo). Pull Request 19625
Update RStudio IT (thanks to @afgane). Pull Request 19711
More user feedback in FormRulesEdit (for Apply Rules tool) (thanks to @jmchilton). Pull Request 19827
Handle directories with percents directories with export_remote.xml (thanks to @jmchilton). Pull Request 19865
Add basic support for icons in tools (thanks to @davelopez). Pull Request 19850
Data-source tool for DICED database (https://diced.lerner.ccf.org/) added (thanks to @jaidevjoshi83). Pull Request 19689
RStudio IT updates to work on .org (thanks to @afgane). Pull Request 19924
Also chown R lib in RStudio BioC tool (thanks to @natefoo). Pull Request 20025
IT Activity Panel (thanks to @dannon). Pull Request 19996
Add missing tool test file (thanks to @jmchilton). Pull Request 19763
Please see the full 25.0 release notes for more details.
To stay up to date with Galaxy’s progress, watch our screencasts; visit our community Hub; and follow us on Bluesky, Mastodon, and LinkedIn.
You can always chat with us on Matrix.
Thanks for using Galaxy!